Fractal Project Sharing¶
The Fractal framework, developed at the BioVisionCenter, allows users to build complex image processing workflows and run them on Terabytes of high-dimensional microscopy data.
With more people using Fractal and collaborating on analysis workflows, we wanted to bring more collaboration features to our users. As a first such feature, we have recently added project sharing functionality to Fractal with the 2.18 Fractal server update.
Project sharing allows users within a given Fractal server to collaborate granularly on their projects. Users can give access for collaborators to see their projects, stream the processed image data or collaborate on building and running workflows.
The ability for project sharing comes together with an improved fractal-data service. Fractal-data allows authenticated streaming of OME-Zarrs to viewers like ViZarr and analysis environments like the Fractal feature explorer. As part of this effort, we have simplified the authorization logic to follow project directories & dataset Zarr directories. This allowed us to add support for data streaming of any datasets in shared projects: If your colleague processed OME-Zarrs in a project shared with you, you now also have access to visualise the images directly in a web viewer like ViZarr or browse the measurements in the Fractal feature explorer.
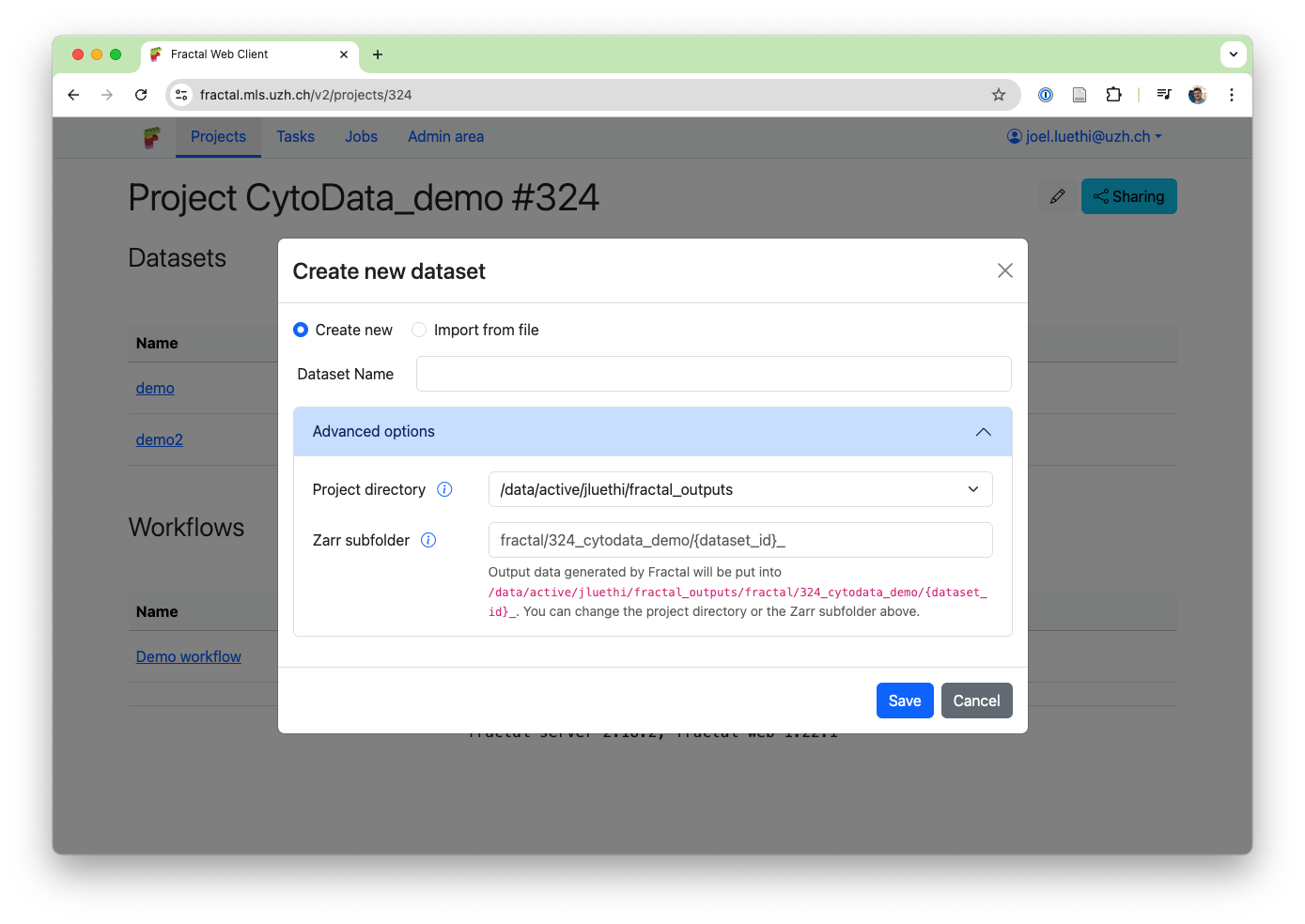
To enable these new data streaming services, we've revamped the project directory logic: Fractal used to default to put your OME-Zarrs in your (single) project directory. With the new update, we now enforce that all Fractal output go into a project directory. This both simplified the dataset creation options (users don't need to know the base paths to the zarr directory anymore to modify the target location of their dataset zarr directories) and ensures that we can safely use project directories for streaming authentication. To handle complex use-cases, we now support multiple project directories for a single user.
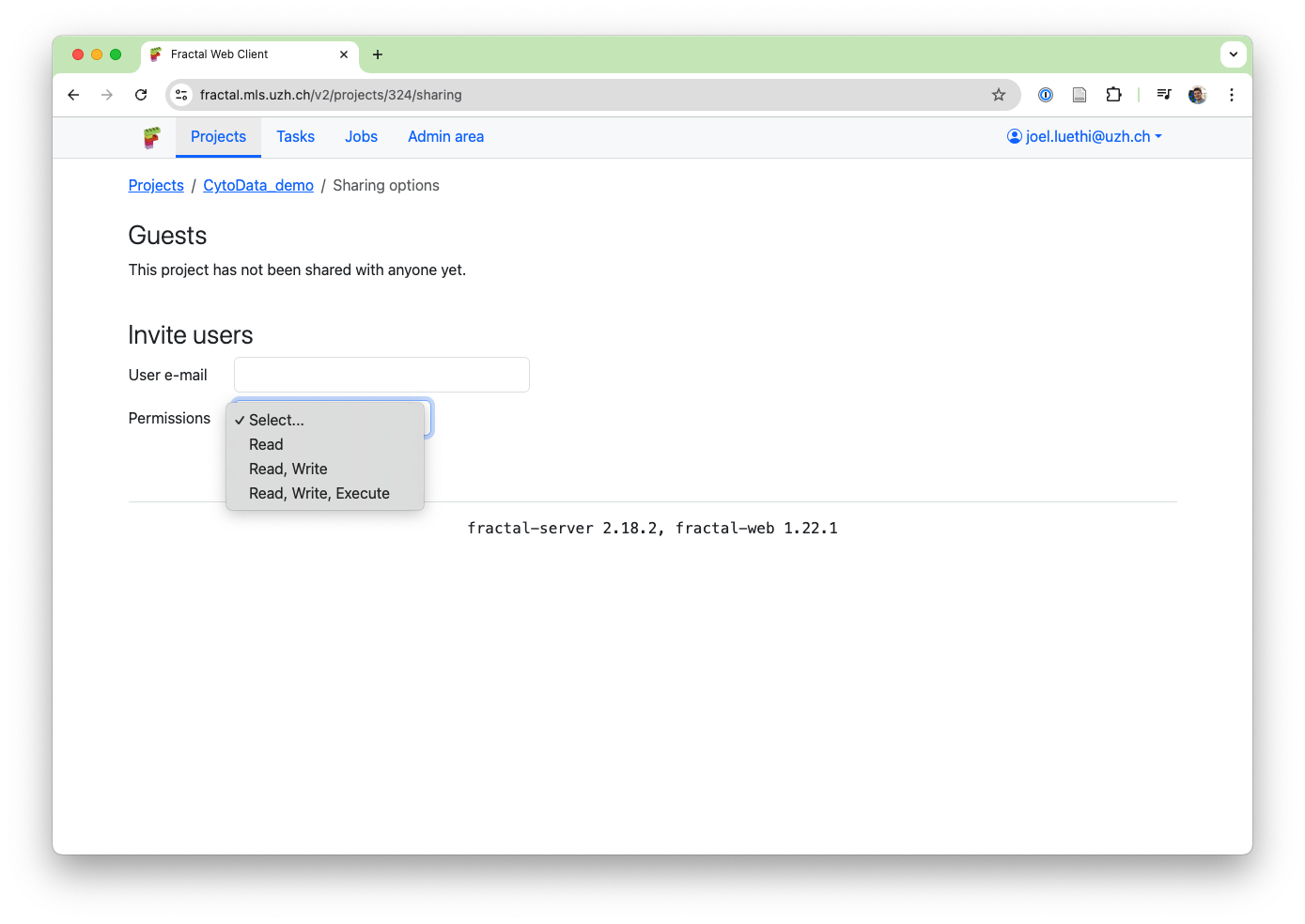
We wanted to support a variety of use-cases for project sharing: from easily showing images and quantification results to a colleague in the lab or collaborating on creating workflows, to members of core facilities supporting users by setting up initial workflows for them or helping them with existing workflows in their projects. To enable this, project sharing permissions are granular: - Share a project in read-only mode to gives access to view the workflows and stream the OME-Zarrs (see above): This ensures your collaborator can see how you processed your data and can see the data itself. - Share a project in read + write mode to allow editing workflows: Want your collaborator to help with workflow parameters? This gives them access to modify your workflow. - Share a project in read + write + execute mode: This gives full access to your collaborator. On top of everything above, they can now also run workflows from the shared project. Share workflow execution comes with some limitations depending on your deployments: Both users need to have data access within the linux cluster environment to modify the same files. In ssh-slurm deployments (like the UZH Science Cluster & UZH Science Cloud servers), this typically holds for users in the same research group, but not across research groups. In sudo-slurm deployments, this depends on configurations (please contact an admin for questions).
You can follow along with our ongoing Fractal developments by keeping an eye on this newly created news section on the Fractal page or by following the development process on our Github repositories.